Genome science reveals patterns of bTB transmission

The five-year study (2014-2018) investigated the effects of selective culling and vaccination on bTB prevalence in badgers, increasing the understanding of badger behaviour and ecology.
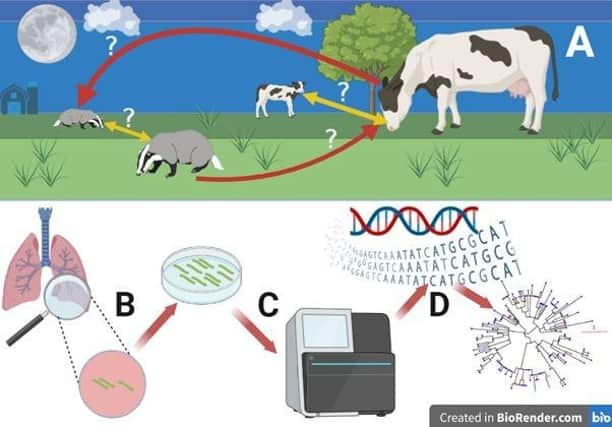
Now, scientists from the Agri-Food and Biosciences Institute (AFBI), funded by DAERA and collaborating with the Universities of Edinburgh, Glasgow and Georgia, have published a new study, which investigates disease transmission within and between cattle and badger populations using whole-genome sequencing of the bacteria that cause the disease. In the course of the study, they generated 162 gigabytes of genetic data from ~600 isolates of the bacterium that causes bTB – Mycobacterium bovis.
Advertisement
Advertisement
This high-resolution genomic data allowed them to construct family trees of the bacteria isolated from cattle and badgers and to infer the likely relative importance of the various routes of transmission (cattle-to-cattle, badger-to-badger, cattle-to-badger and badger-to-cattle)
The study found that, in the TVR zone, cattle-to-cattle transmission was by far the most common form of disease spread observed, while badger-to-badger transmission was undetected. However, signals of inter-species transmission (both cattle-to-badger and badger-to-cattle) were detected as both hosts swapped closely related strains of bacteria. However, while being highly variable, cattle-to-badger transmission appeared to be considerably more common than badger-to-cattle transmission.
The researchers also studied the genetics of the badger population in the TVR zone, finding that over the period of the study, badgers remained highly territorial with closely related badgers tending to stay physically close in the landscape. This being the case, if badgers were commonly transmitting bTB, one might expect closely related badgers to share closely related pathogen strains. The researchers found this not to be the case.
The sum of the evidence suggests that cattle were playing the major role in the spread of disease in the TVR zone, with badgers involved, but to a lesser extent. One important observation, however, needs to be made: While badger-to-cattle transmission was detected less commonly, once it occurs, onward within- and between-herd cattle-to-cattle transmission can amplify such initial seeding events to have a much greater impact. Because the pathogen has been found to mutate incredibly slowly such amplification effects cannot be detected by genomic methods, such as deployed here. However, it remains plausible that a trickle of infection from badgers-to-cattle could still have an outsized effect.
Advertisement
Advertisement
The TVR zone was chosen for intervention by DAERA as it was an area of high badger and cattle density which had a high level of bTB in those cattle herds. The area had also experienced a largely unexplained rise in bTB incidence in 2011-2012. A signal of this rise was clear in the pathogen genome data; pathogen genetic diversity increased markedly in the years preceding the TVR study. Such findings are context-dependent, and it may not be possible to extrapolate across the wider landscape – indeed, transmission dynamics may vary in different locations. Furthermore, the intervention itself of selective badger culling and vaccination may have affected transmission dynamics in a way that the researchers couldn’t observe.